The Omicron differs structurally from all other variants that have been known, and compared to the wild-type Wuhan-Hu-1 strain, 32 out of 50 mutations are located on the spike protein (Figure 1). Recent studies have begun to shed light on the features of the highly mutated virus that have enabled it to evade the immune defenses to viral escape from antibody responses in convalescent or vaccinated individuals. Many of these mutations or deletions affect the N-terminal domain (NTD) and receptor-binding domain (RBD) of the spike protein, which may in turn increase binding to ACE2 while evading antibody recognition.
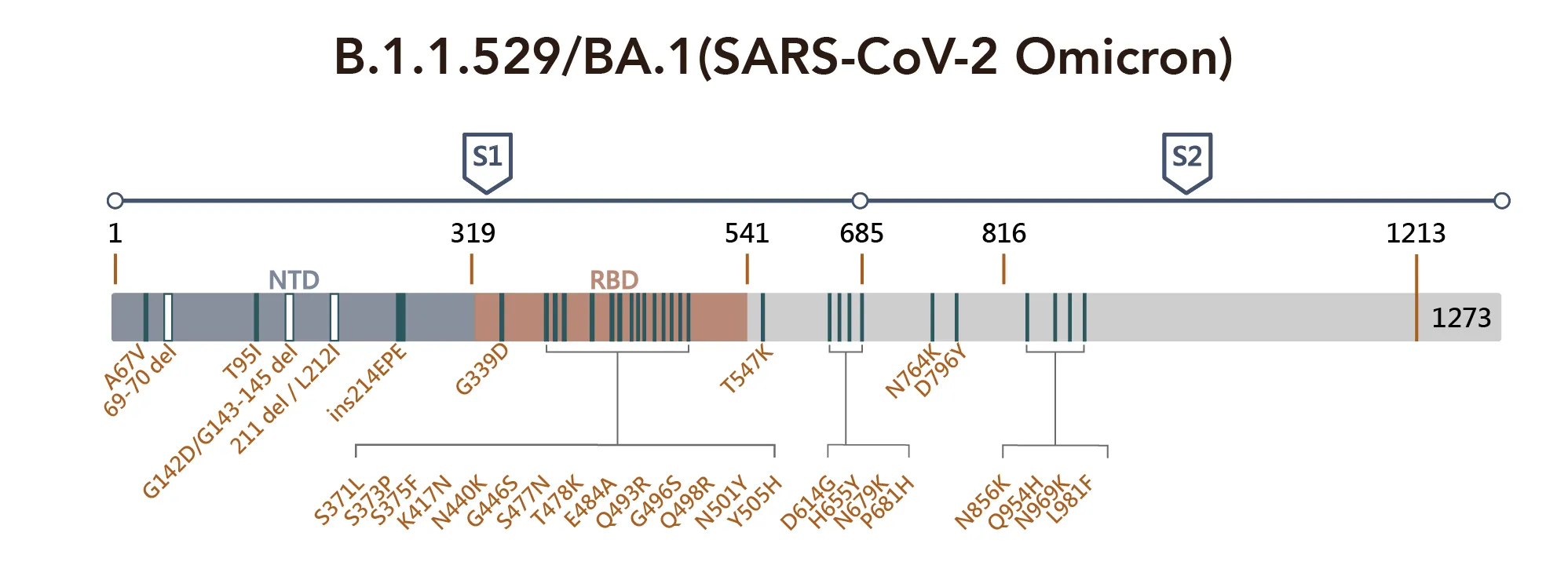
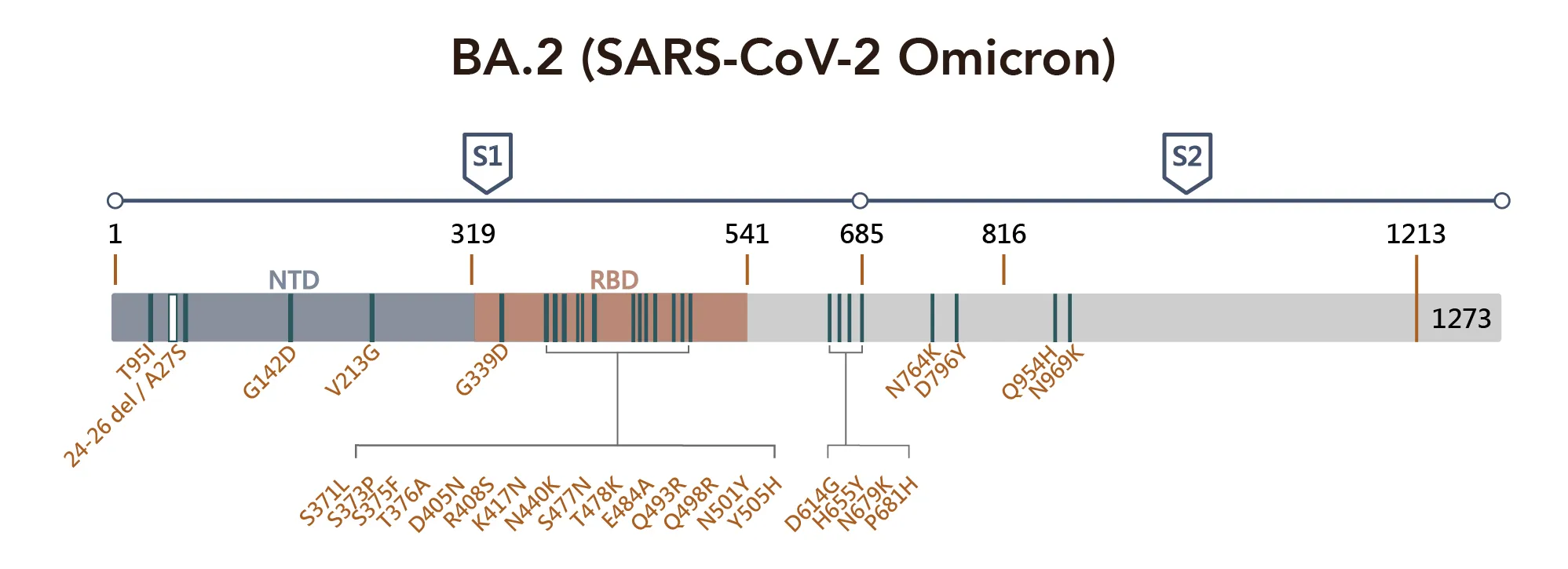
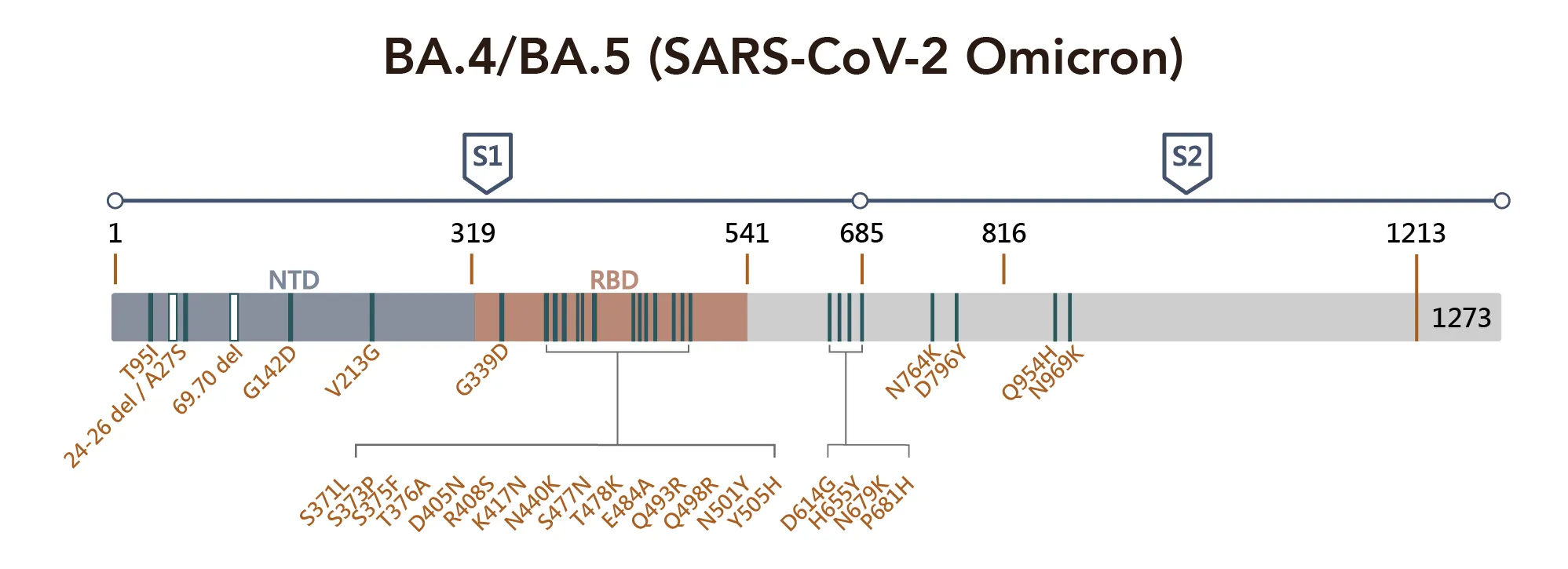
Figure 1. SARS-CoV-2 Omicron Variant Mutation Sites on Spike Proteina
Critical sequence alterations confer emerging variants a survival advantage under the selection pressure of immune surveillance. For example, A67V, T95I, and G142D alter the local conformation of NTD, which most likely disrupts the binding of NTD targeting antibodies. Similarly, deletions in amino acids 241–243 and 246–252 have been found to affect the network of NTD loops in strains with high transmissibility. SARS-CoV-2 RBD has a highly flexible region between residue 475 and 485; the position S477 shows the highest flexibility among them. The double mutation: S477N and T478K have been reported to not only increase binding to human ACE2 but also impact the overall protein expression levels, promoting the virus yield and infectivity, though it does not pose a problem to antibody binding.
Several mutations found in the RBD are found in other variants of concern and may increase the viral infectivity, such as D614G, N501Y, and K417N. While the K417N mutation acts together with N501Y, some neutralizing antibody groups become fully abolished. Notably, residue 484 has mutated into various amino acids under the pressure of SARS-CoV-2 convalescent sera, and mutation at this site can cause immune resistance to different convalescent sera. Diversity on 484E implies that the location of the recognition "hotspot" of polyclonal antibodies and the time that amino acid changes are found in emerging variants consistently exhibit broad resistance.
All three sublineages of the Omicron harbor the E484A mutation, further supporting that this mutation can somewhat explain the decreased susceptibility to neutralization by convalescent sera. Again the D614G and other four substitutions: T547K, H655Y, N679K, and P681H, may facilitate rigidity or flexibility of spike protein conformational switches, resulting in increased furin cleavage, and consequently enhancing cell tropism and viral transmissibility. Mutations such as N764K, D796Y, N856K, Q954H, N969K, and L981F all occur in the S2 subunit, which is the region that is responsible for their pre-and-post-fusion structures. In addition, this region is supposed to increase infectivity by facilitating the spike-ACE2 fusion.
Read More
Tracing Back the Mysterious Ancestry of SARS-CoV-2 Omicron Variant
Does Omicron Variant Cause Milder Symptoms?
